Chapter 3
Introduction to genetic testing
What you’ll learn in this chapter
In this chapter, we’ll cover:
- The idea that biology is about probability rather than a given outcome;
- How genetic testing works;
- Why and how genetic testing might be helpful or unhelpful;
- Guidelines for genetic testing; and
- Questions you can ask yourself when considering genetic testing.
Biology is probability.
Probability is prediction and potential, not perfection.
Biology rarely gives us a definitive, simple answer to questions like:
“What is the best diet?”
or
“What is the best exercise routine?”
or
“When exactly will I die, and how?”
So any genetic testing service (along with any book, website, or coaching service) that promises to give you a perfect nutrition plan or workout regime, or exactly predict your health risks, is being misleading.
(And scientifically dodgy.)
There will almost never be a “perfect solution” for anything related to biology.
As we like to say around Precision Nutrition:
“Progress, not perfection.”
It’s almost impossible to control all the factors involved in a complex system.
Plus, knowledge and development are incremental. We learn bit by bit. Change bit by bit. Grow bit by bit.
Genetic information can tell us how we might change (if we wanted to), and what the payoff from that change might be.
Let’s imagine that based on your genetic tests, you knew exactly how likely it was that you would get a particular disease, and exactly how that disease might progress.
(Just to be clear, we’re definitely not there yet. It’s imagination time only.)
For instance, let’s say that you know from genetic testing that you have a 70% probability of dying from Alzheimer’s disease by age 70.
Now let’s say that there is also a medication available. This medication will bring your risk from 70% down to 30%. And it works in 90% of people.
Would you take it?
Let’s think about that.
No medication: 70% chance of dying at 70 from a known disease.
Medication: 30% chance of dying at 70… but you may be in the 10% for whom it doesn’t work.
Many people would probably figure OK, I am probably more likely to be in the 90% of medication responders, and I like that 30% is much less of a chance than 70%, so let’s do it.
What if the medication only brought your risk down to 60%?
Or if it only worked in 30% of people?
What then?
Here’s another example.
We know that many breast and ovarian cancers are related to mutations in the BRCA1 and/or BRCA2 genes. Data suggest that in healthy 30-year old carriers of these mutations, removing ovaries may add 0.2 to 1.8 years in life expectancy, and a mastectomy may add 0.6 to 2.1 years.
Is that enough certainty to book surgery if you know you carry those mutations?
Of course, there’s no right answer.
Some people might grab for any chance, and take the medication, or have surgery. Other people might say Eh, not good enough odds for me, and not take those chances.
The point is:
Unless a mutation has taken you out of the game right off the bat, there’s almost never 100 – 100 – 100.
As in: We know with 100% certainty that in 100% of cases, this genetic risk will 100% lead to this outcome.
Sometimes we don’t even know with 50% certainty… or 10% certainty… or at all.
At best, we can only make informed guesses.
Proceed with caution and critical thought.
Now that you know from Chapter 2 the basics of how genes work, remember a few key ideas:
- Genetic interactions are vast and complex, and we don’t have most of them mapped.
- Genetics isn’t the whole story. Many physiological interactions aren’t genetic.
- This domain of research is still very new.
- Mutations and alterations to our genetic code happen all the time. Sometimes they matter. Sometimes not.
- We still can’t make strong predictions or recommendations about most things.
Keep these concepts in mind, and wear your “skeptical scientist” hat.
Genetic testing is easier than ever. What does that mean?
In the last few decades, technologies that measure DNA and RNA have developed rapidly, becoming cheaper, faster, and more accessible.
- In 2001, sequencing a full human genome (all the DNA in a human cell nucleus) cost about $100 million US.
- In 2011, it cost about $10,000 US.
- In 2016, two members of our PN team got their full genome sequenced for $1,000 US each.
Still too much money?
For only $200 US, you can buy a 23andMe kit and get hundreds of thousands of your genetic variations tested without ever leaving your house. Just spit in a tube and mail it away for analysis — no fancy lab coat required.
The graphic below compares when various genetic sequencing technologies were introduced to how much analytic power those technologies have.
When more material can pass through the process, accuracy goes up and costs go down. Researchers can now do high-fidelity sequencing on the entire human genome at a cost that even projects that survive on grants can afford.
Note that the Y axis increases logarithmically (10, 100, 1,000, 10,000, etc.). What looks like a small step on this graph’s Y axis is actually a huge step forward: The Illumina HighSeq X Ten sequencer has 10,000 times the throughput as the Sequence Analyzer Illumina created just ten years before.
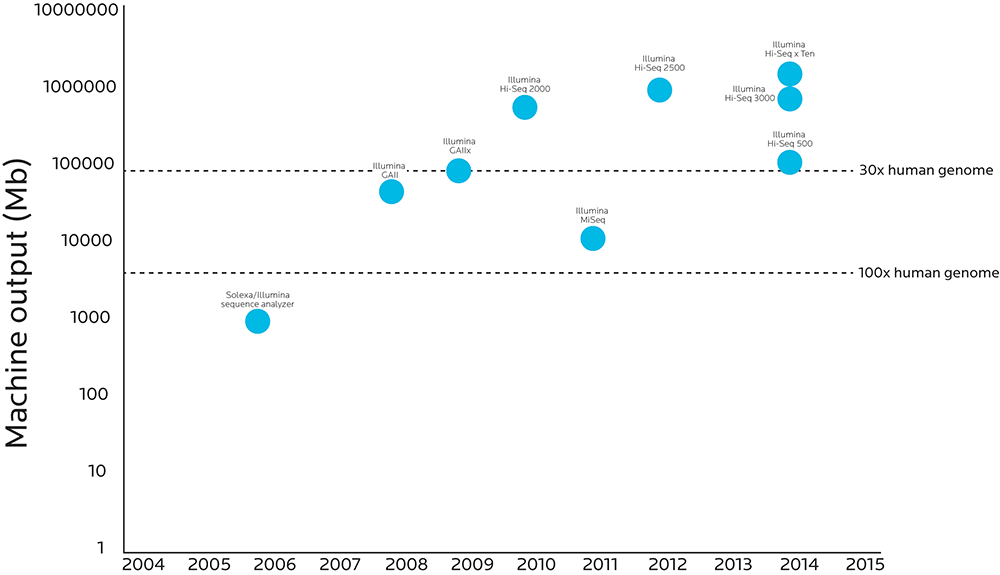
We can look at the genetic code with more precision and accuracy than ever before.
This means we also need to consider how to deal with all the data we generate, which has to be compiled, converted, and stored. We must then analyze the results and balance any clinical advice or interpretation that we give.
This means that each stage of the genetic testing process is significant, such as:
- collecting, transporting and storing a sample properly;
- “reading” a sample correctly;
- analyzing the sample;
- storing the data; and
- interpreting the data — deciding what it all means, and what to do next.
Generating so much data means that we have to think about where to put it, how to understand it, and what to eventually do with it.
We can think about it in terms of bioinformatics — combining technical domains like computer science, statistics, mathematics, and engineering to analyze and interpret the data we’ve collected.
We can also think of it in terms of behavior change and genetic counseling — what we might do with any data or insight we gain.
We have to be careful about making too many assumptions about this process.
There is a lot that we don’t yet know. A lot of things can go wrong with this complex process. This affects the quality of the data we get, and the conclusions that we can draw.
For example:
- Many diseases or health conditions with a strong genetic basis are rare or less common. Testing for these genetic variants may not apply to most people.
- Many diseases and health conditions are complex. There may be a genetic contribution, but it may be from several interacting genes, or even regulatory pathways that don’t involve genes. And environmental or lifestyle choices may affect the outcome more anyway.
- Other outcomes, such as athletic performance, are also complex. For instance, is there a “sprinter gene”? (Spoiler: No.) What if you have a certain set of genes that give you muscularity and power, but not the set of genes that give you the motivation to show up in the morning for a workout?
- Data are limited. There may not be a lot of research about a particular gene, or its relationship to health and function.
- We don’t always know what to do about test results. In fact, research suggests that just knowing what’s in our DNA rarely changes our behavior.
- The technologies for testing, analyzing, and interpreting genetic materials continue to evolve and change quickly.
- Genetic testing services — perhaps driven by commercial interests, or patient advocacy groups — are not always completely honest about what their tests can and cannot do. They may market their products as more useful or revealing than they really are.
To understand this better, let’s look more closely at the process of genetic testing.
What does clinical genetic testing involve?
In general, clinical genetic tests:
- analyze DNA for specific genetic variants (SNPs), that code for gene products such as enzymes or other proteins;
- look for variations that are related to disease or health (and, increasingly, athletic capacity);
- focus on a particular population (such as an ethnic group with a higher risk of a specific hereditary disease); and
- are aimed at producing information that patients can somehow use — for instance, to lower their risk of a disease, to choose the right medication, or adjust their nutritional regime.
Some genetic tests (such as 23andMe) also include ancestry and ethnic heritage. We’ll look at the importance of ancestry in Chapter 5.
An assay is a method for determining the presence or quantity of a particular component, or a method to analyze or quantify a particular substance in a sample.
A genetic test is a laboratory assay specifically for clinical testing. It identifies specific genotype(s) to diagnose a specific disease in a specific group of people for a specific purpose. Genetic testing is very targeted compared to a genetic assay, which may be scanned, or may be targeted.
An open-ended assay looks for anything of interest, like scanning a landscape to see what pops out.
A closed assay identifies beforehand what it’s seeking, such as a particular mutation or another variant.
Genetic tests might be used for:
- disease diagnosis;
- health risk predictions;
- carrier testing (in other words, if you carry a gene variant that isn’t active in you, but that you may pass along to your offspring);
- prenatal testing, to identify particular conditions in fetuses;
- newborn screening, done soon after birth to look for potential diseases;
- pharmacogenomic testing, to explore how you might respond to a particular medication; or
- research — either for a specific purpose, or for general investigation.
Increasingly, some people are also exploring genetic testing for nutritional needs and athletic performance.
For instance, genetic tests can currently help us explore such traits as:
- how quickly you process caffeine;
- how your body processes vitamin D;
- how much inflammation you’re likely to have (for instance, by testing for C-reactive protein, a marker of inflammation);
- how well you may recover from exercise; or
- what your body weight range is more likely to be (for instance, whether you are more likely to have a higher BMI).
Yet few of these are definitive or clear.
Test types
You might think that a genetic test would look at the entire genome, but this is rarely the case. (We’ll talk more in upcoming chapters about studying entire genomes, and why most commercially-available tests don’t do it.)
- Molecular genetic tests look at the smallest “chunks” of DNA — perhaps a single gene or short pieces of DNA — usually looking for a specific variation or mutation. An example of this might be testing for the cystic fibrosis variant or the BRCA1/2 mutations that are linked to breast and ovarian cancers.
- Chromosomal genetic tests look at longer pieces of DNA, such as whole chromosomes, to look for larger-scale genetic changes (such as an extra chromosome copy). An example of this might be testing for Down syndrome.
- Biochemical tests look at how much of a certain protein we have, or how active that protein is. Here, the test doesn’t look at the DNA, but rather the protein that it might be coding for. By looking at differences in the proteins, testers can speculate about genetic variations. An example of this might be a c-reactive protein (CRP) immunoassay; CRP is a protein marker of inflammation.
- Genetic sequencing involves “reading” a strand of DNA by looking at its nucleotides, one by one. The first sequencing of a full genetic code was done on a simple virus (known as Phi X 174) in 1977; the human genome sequence (or at least, 90% of it) was published in 2001. A test that reads the entire genome is known as whole-genome sequencing.
Again, commercial tests don’t usually sequence entire genomes.
In fact, usually the opposite is true: Most commercially available genetic tests examine single-nucleotide polymorphisms (SNPs), a variation in a single nucleotide (e.g., having a cytosine, or C, where there’s normally an adenine, or A). You’ll remember from Chapter 2 that small substitutions like this are one way that we can get genetic variation.
SNPs and noncoding DNA
The testing service 23andMe’s genotyping test covers about 600,000 SNPs of the potentially 3 billion SNPs that each person has… or just 0.02% of the genome. Even the most comprehensive genotyping services covers less than 0.15% of the genome.
Of those SNPs, 98.6% are in noncoding DNA, which means they are stretches of DNA that aren’t transcribed into mRNA to make proteins. (This is what people used to call “junk DNA” until they realized that it wasn’t junk. Sure, some of it includes leftover odds and ends from our evolutionary history, but a lot of it has a purpose.)
Noncoding DNA includes a lot of different things. Much like after family get-togethers when you end up with lots of leftovers and macaroni-salad-encrusted Tupperware, our genome contains lots of leftover bits from millions of years of our evolutionary process.
For instance, we have:
- Pseudogenes: genes that used to be active but aren’t any more.
- Proviruses: There is a class of viruses, known as retroviruses, that transcribe their genome and insert it into the genome of the cells they infect. This genetic material is known as a provirus. HIV is probably the best-known example of a retrovirus, but there are many others. Because the process by which this happens (known as reverse transcription) is relatively unstable and prone to error, many of these retroviral sequences are inactive. Inactivated retroviral sequences make up a surprisingly large proportion of our genome — about 10%.
- Regulatory sequences: parts of DNA that act like a volume knob on gene expression: They can “turn it up” (increase the genetic expression of a certain protein), “turn it down” (decrease expression), or even “turn it off” completely (prevent expression).
So, genetic testing services may test for functional SNPs, or they may test for SNPs found in noncoding regions.
On one hand, we can get a lot of useful information from SNPs, such as predictions about our ancestry, or well-known associations with certain heritable traits or diseases.
On the other hand, using SNPs depends on having known associations, or identifying regions and points of common variation. We must already have identified and interpreted these SNPs — where they are, how common they are, and sometimes what they do, to what degree, and in which population. This is much like only getting information from books you’ve already read.
We also can’t see copy number variation (again, how many copies of a particular DNA sequence you have) nor translocation mutations (where a gene moves from one chromosome to another, or from one part of a chromosome to another). Both of these can affect our phenotype, but simply knowing SNP markers won’t tell us anything about their effects.
Thus, there are tradeoffs.
Genome-wide association studies (GWAS)
While commercial tests rarely look at whole genomes, laboratory research that is looking for the relationship between particular gene variants and outcomes such as health problems or biological processes might do genome-wide association studies (GWAS).
With most GWAS, participants are usually picked based on some characteristic that they share, such as having high blood pressure or osteoporosis. They are then compared to a control group without that characteristic to see if the test group shares any genetic variants, or somehow differs from the control group.
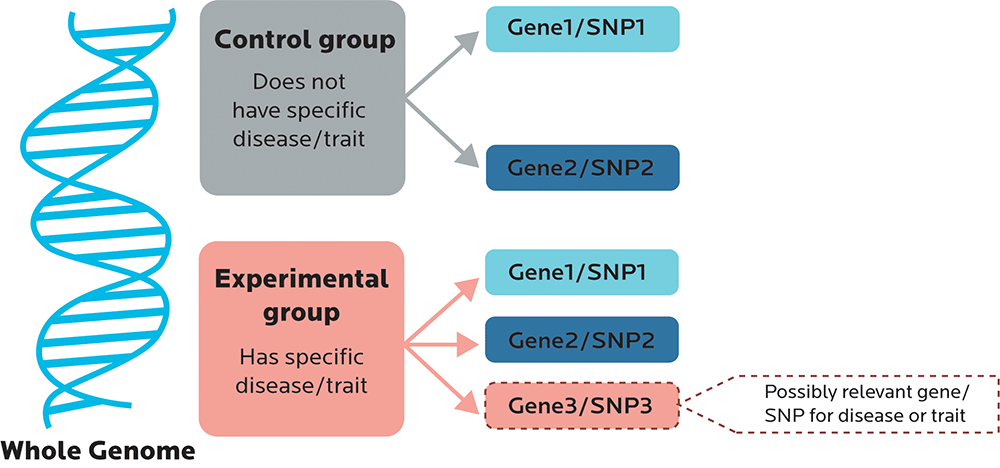
A GWAS is usually a broad scan.
- Sometimes researchers are looking to confirm that Gene Variant X is, indeed, significant.
- Or maybe they’re looking to find novel loci — in other words, new SNPs or other variants such as haplotypes (which we’ll look at in Chapter 5) that are associated with the characteristic.
- They’re also looking to see whether the SNPs have predictive value: In other words, does having this SNP make it significantly more or less likely that you will have a certain trait or other outcome? Or is the relationship between the SNP and the outcome just kinda random?
Often, the data that commercial genetic testing services use to look for certain SNPs come from laboratory GWAS that have already identified those SNPs as significant.
Commercial direct-to-consumer testing vs. experimental lab testing
While we can now access many of the same scientific methods and tools that researchers might use in high-end experimental labs, there are a few key differences between them.
Partial vs. full-genome testing
At the time of writing this book, no one currently offers a commercial full-genome testing service. (However, we did get our co-author Alaina’s genome sequenced in a private lab. More on that later.)
Right now, whole-genome sequencing is expensive. However, as the price drops and researchers make discoveries about the intricacies of the human genome, whole-genome sequencing will probably become more and more common. Eventually your whole-genome sequence will just be a normal part of your medical records.
It may seem like a commercial test is only offering you a half-assed version of a “real” full-genome test, but when you know what parts of the genome you need to look at, there’s no reason to sequence the entire genome.
It’s faster, cheaper, and easier to look at only the regions of interest, so you aren’t looking for a needle in a haystack. Instead, you already know where the needle is; you just need to know how long it is.
The only challenge is that research hasn’t uncovered all the different relationships between our genes and our genetic expression. The scientific community has only scratched the surface.
So:
- Commercially available testing services can tell us things about ourselves based on what research knows today. That’s great if we’re looking for a specific, well-researched genetic factor and belong to a population that’s been extensively studied.
- These services are really only telling us a small fraction of all there is to know about ourselves. That’s challenging if we’re looking for the best way to eat, or exercise, or live to 120.
What is the process of genetic testing?
Although there are various types of genetic tests, here’s a general overview of how most of them work.
Step 1: Collect a sample.
First, the test requires some type of sample material to analyze. In theory, this can be any type of biological material, but is most often:
- blood (though red blood cells would have to be removed);
- buccal cells (aka a swab of skin cells from the inside of the cheek);
- amniotic fluid (the fluid that surrounds a fetus in the womb); or
- hair (specifically, the cells from the follicle).
Criminal investigations using DNA as evidence may also use “discarded DNA”, such as material left behind on coffee cups, straws, or cigarette butts.
Step 2: Prepare the sample.
Once the sample is collected and sent to the lab for analysis, we have to somehow get the DNA out of its container, the cell, and read it.
Step 2A: Get stuff out of cells.
This means that first, the cells have to be ruptured, a process known as lysing (from the ancient Greek lusis, or loosening).
Because cell walls are lipid-based, we have to use some kind of surfactant or detergent, much like dunking the cells in dish soap.
Some processes may also use a base such as sodium hydroxide (NaOH) along with the surfactant sodium dodecyl (lauryl) sulfate (SDS). (The combination of a base like NaOH and a surfactant is known as alkaline lysis, and it was first described in 1979 as a way to get DNA out of bacteria.)
Once the DNA is out of the cell, testers break up proteins using proteases (protein-degrading enzymes) and break up RNA by using RNases (same idea). Without this step, the sample can degrade rapidly and cause sampling errors.
Luckily, DNA is remarkably stable outside of the cell, much more than RNA or protein. This is why we can do DNA matching days or even years after a sample has been left. In fact, ancient DNA can be extracted and analyzed from samples thousands of years old.
Step 2B: Separate the solution to get what you want.
Now that you’ve got a solution of cell crud, add a concentrated salt solution to make all the bits and chunks clump together.
You need to pick apart the crud from the DNA, so the next step is centrifuging, which spins the mix in a wheel to separate things out (much like that spinning wheel ride at the county fair that separates you from your stomach contents). Solid chunks are flung to the bottom of the test sample tube, and the DNA is left behind, dissolved in the solution.
Next, you want to tidy and purify the DNA from all the stuff you used to get it out of the cell. Do one of the following:
- Use ethanol (the same alcohol as in your martini) or isopropanol (aka rubbing alcohol, definitely not in your martini) to come out of the solution (precipitate it). DNA won’t dissolve in this, so it clumps when you centrifuge it. If this clump is clear it’s pure DNA. If there’s still some non-DNA, the clump is white.
- Use phenol-chloroform: Denature proteins with phenol; whisk nucleic acids away with chloroform. This works sort of like making a salad dressing with oil and vinegar.
- Use something solid, like silica, to which nucleic acids will bind. This works sort of like dumping sawdust on spilt liquid. Most labs that have money use silica bound to a membrane. This is faster and it gives us clean DNA.
- Use a protease to break up and cleave off cellular and histone proteins bound to the DNA.
Step 2C: Amplify the DNA.
Let’s say you’re trying to listen to music, but you’re somewhere with lots of background noise. So what do you do? You turn up your music — in other words, you amplify it.
The same thing happens in DNA analysis — you often have to amplify it and make a lot more copies of the strand of DNA in order to make sure you get a good “signal” to test. This is most often done using the polymerase chain reaction process, or PCR.
The upside of this process is that after several rounds of amplification, you have a nice big sample — more DNA than you know what to do with. The downside is that if there’s contamination, that’s amplified too.
In tightly-controlled experimental studies, researchers control for contamination. The same may not be true of commercial tests.
Fun factoid!
Legend has it that the inventor of PCR, the Nobel Prize-winning American biochemist Karey Mullis, got the idea for PCR while on an acid trip in 1983.
Also, he believes he was visited by an extraterrestrial bioluminescent raccoon and denies anthropogenic climate change.
Goes to show that winning a Nobel Prize and transforming molecular biology doesn’t mean you aren’t also a crackpot.
Step 3: Analysis
Once you have a nice big pile of DNA, you can start to “read” (i.e., sequence) and analyze it.
“Reading” DNA isn’t as simple as reading a book from start to finish. It’s more like this:
- Take a book.
- Open the book to a randomly-chosen page.
- Start reading at a randomly-chosen word.
- Read a few letters, or a word, or a phrase. Maybe a sentence.
- Close the book.
- Open the book to another randomly-chosen page.
- Read a few more randomly-chosen bits.
- And so on.
- Do that about a zillion times until you understand what the book says.
In real-life terms, that means that sequencing creates a bunch of data that need to be put together into an order that makes sense. We do this with complex computing methods and algorithms, using bioinformatics techniques to identify, understand, and analyze the raw data.
Step 4: Interpretation
Finally, once you have your data, and it’s been read and analyzed, you can decide what your findings mean. For example, did your sample have the variant you’re looking for?
What do you need to know about this process?
Obviously, you could write a Ph.D. thesis on a single aspect of genetic testing alone.
But here are a few key points:
- There are several types of tests, depending on what you want to find.
- Different tests use — and look for — different amounts of genetic material.
- There are several methods for testing.
- There are several steps in the process, each with the potential for variation or mistakes.
- It’s complicated.
How do you know if genetic testing is useful or valuable?
Why test?
Defining “useful”, “beneficial”, or “valuable” depends a lot on what we are seeking from a genetic test.
For instance:
- A researcher may be interested in the test methods themselves, and how they advance basic science.
- A computational biologist may be interested in new forms of data analysis.
- A clinician may want to explore associations between particular genetic variants and disease risk.
- A family doctor may want to know how to advise their patients about medications, family planning, or lifestyle choices.
- A genealogist may be curious about ancestry.
- An evolutionary anthropologist may want to know about interesting features of specific populations, their origins, or how a trait spread across a population.
- A sports scientist may want to know how to select or train athletes for optimal performance.
- A family lawyer settling a patrimony case (or a sleazy TV talk show host doing an “OMG! Who’s Your Baby Daddy?!” episode) may want to know which kids are the genetic offspring of a particular parent.
For many people, genetic testing is a way to explore their risk of disease or health conditions. Yet there is much we still don’t know about genetic contributions to disease, and how we might use information about our DNA to either treat specific diseases, or improve our odds of staying healthy and fit.
In general, one definition of a genetic test is that it has a purpose.
Researchers are trying to find something, even if they aren’t always exactly sure what. And there is some reason for them to look, such as advising people about reproduction or disease risk.
A cautionary tale about the science of testing
In 2006, some scientists published their findings about a method for how to use gene expression profiles to personalize chemotherapy and other cancer drug regimes.
When a second group of scientists tried to replicate these findings, they found that, as they said, “poor documentation hid many simple errors that undermined the approach.” This is a polite sciencey way of saying that the first team might have fibbed a bit.
However, these problematic profiles were used anyway to direct patient therapy in clinical trials at Duke University in 2007. When the second group of scientists protested the poor science, trials were suspended in 2009, then re-started, and then finally ended in 2010.
The original 2006 study was retracted.
The initial exploration held exciting promise: What if cancer patients could have their treatment individualized, based on their genetic profile?
Yet the scientific reality did not hold up under scrutiny.
That isn’t to say that this couldn’t happen one day. It very likely could.
The point is that the science is young, and all results must be reproduced reliably before we can have confidence in them, or use them to guide our decisions.
Establishing guidelines for genetic testing
The researchers who protested the Duke study suggested that all genetic tests share the following:
- the raw data, such as the reads produced by a sequencer; the called genotyping data in the file you can download from 23andMe; or the images taken of gels;

- the code used to derive the results from the raw data (in other words, how software and algorithms computed and analyzed the findings);
- evidence of the origin of the raw data so that labels could be checked;
- written descriptions of all the steps in the analysis; and
- how the researchers planned to run the analysis.
Originally, these were meant as guidelines for researchers who wanted to publish their work in scientific journals, but the scientists also suggested that anyone starting a clinical trial that used genetic data to guide treatment should meet these requirements.
In 2004, the Centers for Disease Control and Prevention in the United States established the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative to “establish and test a systematic, evidence-based process for evaluating genetic tests and other applications of genomic technology that are in transition from research to clinical and public health practice.”
The EGAPP wanted to have a broad perspective on genetic testing. The group’s founding members included experts in:
- evidence-based review;
- clinical practice;
- developing clinical guidelines;
- public health;
- laboratory methods;
- genomics;
- epidemiology;
- economics;
- ethics;
- policy; and
- health technology assessment.
EGAPP aimed to help healthcare practitioners and their patients understand some of the results of genetic tests.
After reviewing many genetic tests, they concluded that:
- Only a few had enough evidence to be considered clinically useful.
- Many simply did not have enough evidence yet to help patients and their doctors make decisions about health risks and treatment.
In 2010, the United States Institute of Medicine’s (IOM) Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials met to discuss the challenges of genetic testing.
The group concluded, based on reviewing how genetic testing was being used, that “problems were more widespread and severe than we knew”.
For instance, many tests could not be reproduced accurately; many clinical trials based on genetic data were called into question. Along with more complex problems like statistical analysis, researchers were making basic mistakes like mislabelling data.
Clearly, a better system for scientific rigor was needed.
This reminds us to always be critical and careful of grandiose claims for genetic testing.
Genetic testing holds exciting promise, and may help us make incredible breakthroughs in human understanding.
This promise must be tempered by thoughtful and careful review.
How can you decide whether a genetic test makes sense?
If you’re not a scientist, it can be hard to figure out whether a particular genetic test is helpful.
Here are two ways to think about the answer: a simple 4-question rubric from us, and a more complex checklist from the ACCE.
Keeping it simple: 4 questions to ask about genetic testing
Is this particular test:
- Descriptive: Does it tell me something about the person being tested?
- Diagnostic: Does it allow me (or a medical professional) to diagnose a problem or characteristic?
- Predictive: Does it allow me to predict some future challenge or occurrence, such as a disease or health risk later in life?
- Prescriptive: Does it tell me what to do next, or in the future?
More complex: ACCE criteria for disease-related genetic testing
If you want to think more deeply about how good or useful a genetic test is, you can use the framework proposed by the ACCE (established by the Centers for Disease Control and Prevention in the United States).
ACCE takes its name from the four criteria for judging the value of a given genetic test:
- Analytical validity:
- How accurately does a given test detect a gene variant or mutation?
- How reliable and repeatable is a given test?
- Are test results “lab significant” or “real world significant”? In other words, if we find anything, does it mean anything?
- Clinical validity:
- What evidence supports the relationship between particular gene variants and risk of disease?
- Which variants in particular are important?
- How great are those risks?
- How have those risks been estimated?
- Clinical utility:
- How useful are these findings for making informed judgments about one’s health and medical treatment?
- What should healthcare practitioners and patients do about genetic test results?
- Ethical, social, and legal issues:
- How might privacy, social wellbeing, or legal status be affected by the results of a genetic test?
- Are patients informed of all their rights and obligations beforehand?
- Would prenatal screening be considered appropriate?
- Who has access to these tests?
To help explore these questions, the ACCE has produced a checklist that can help evaluate particular genetic tests, especially those aimed at finding relationships between gene variants and disease risk.
The ACCE checklist
Test purpose and context
- What is the specific clinical disorder to be studied?
- What are the clinical findings defining this disorder? (In other words, what does the existing clinical evidence tell us about diagnosing this disorder, and its specific features?)
- What is the clinical setting in which the test is to be performed?
- What DNA test(s) are associated with this disorder?
- Does this test include preliminary screening questions? For instance, does the test also ask about family history, or look at overall medical and lifestyle factors?
- Is it a stand-alone test or is it one of a series of tests?
- If this test is part of a series, are all tests done at once, or are some tests done based on the results of previous tests? (e.g., if Test A finds something, then do Test B?)
Analytic validity
- Is the test qualitative or quantitative? For instance, is it based on something self-reported or subjective, or on something numeric or objectively measurable?
- How sensitive is the test analysis? How often is the test positive when a specific mutation is present?
- How specific is the test analysis? How often is the test negative when a specific mutation is not present?
- Is the test method regularly monitored and evaluated for quality control by an external body? In other words, who and what tests the test?
- Have repeated measurements been made on specimens?
- What is the within- and between-laboratory precision? In other words, if the same lab were to run the tests twice, or different labs were to repeat the tests, how close would the test results be? (We’ll look at this more in an upcoming section, when we look at what we found with sending samples to different labs.)
- If appropriate, how is confirmatory testing performed to resolve false positive results in a timely manner?
- What range of patient specimens have been tested?
- How often does the test give a useable result? Or fail to do so?
- How similar are results obtained in multiple laboratories using the same, or different technology?
Clinical validity
- How sensitive is the test for clinical purposes? How often is the test positive when a specific disorder is present?
- How specific is the test for clinical purposes? How often is the test negative when a specific disorder is not present?
- Are there ways to resolve clinical false positives quickly?
- How often is this disorder found using this method?
- Has the test been adequately validated on all populations to which it may be offered?
- How many false positives or negatives does the test produce?
- What are the genotype/phenotype relationships? In other words, does a genetic variant actually do anything noticeable? Can we see or measure that impact? If there’s a gene for a trait, does that trait show up a little, a lot, or not at all?
- What are the genetic, environmental or other modifiers? In other words, how significant is the role of the genetic component compared to other factors, such as lifestyle choices?
Clinical utility
- How does the disease normally progress? Can we actually intervene in that process?
- How will the test results affect patient care?
- Are there other diagnostic tests available to confirm the results?
- What can be done about these results, if anything? For instance, are there medications, actions, or some other measurable benefit to knowing these genetic test results?
- If the patient can do something about the results, can they access that? For instance, can they get the medication that may help them, or make any applicable lifestyle changes?
- Is the test being offered to a socially vulnerable population?
- What quality assurance measures are in place?
- What are the results of pilot trials for these specific conditions or diseases?
- Are there known health risks that benefit from follow-up testing / intervention?
- Is testing affordable?
- Are there service providers able to help people understand or take action with the test results?
- Are there evidence-based educational materials that can help patients understand their results?
- Are there informed consent requirements?
- Are there methods for long term monitoring?
- How well does this program work over the long term, and how do we know?
Ethical, social, and legal issues
- When it comes to this particular test, do we need to think about:
- social stigmatization;
- discrimination;
- privacy/confidentiality; and/or
- personal/family social issues?
- Are there legal issues regarding consent, ownership of data and/or samples, patents, licensing, proprietary testing, obligation to disclose, or reporting requirements?
- What safeguards are in place to protect participants?
Other questions to ask about genetic testing
Genetic testing isn’t just about the science or clinical use. There are other factors to think about as well.
How strong or compelling are these results?
Can these results be replicated reliably? (We’ll look at this in an upcoming chapter.)
Are these results a “for sure”, a “maybe”, or “I dunno”?
How high is the risk or probability of a particular outcome? Do you have a 0.5% higher chance of something? 5%? 50%?
What happens if you find a gene variant, but don’t know what it does? Or don’t have any evidence-based strategies for what to do next?
How do these results matter in context?
Are you just a hobbyist who is curious about what’s in your DNA?
Are you someone who’s looking to start a family, and wondering what you might pass along to a child?
Are you someone with a family history of a particular disease, and looking to explore your risk for that disease?
Are you curious about your ethnic ancestry?
Speaking of that…
Do these results align with your genetic and ethnic ancestry?
What group was the genetic research done on?
If you’re ethnically Hmong from Vietnam, or Quechua from Peru, how applicable are genetic studies done (for example) on British Europeans?
What environmental factors could affect these results?
For instance, does a gene you carry only “turn on” if you’re exposed to cigarette smoke, or sunshine, or shift work?
What legal and regulatory factors are involved?
Each jurisdiction may have different rules about what can be tested and shared.
Who can access your genetic information, and for what purpose? For instance, can insurance companies review your genetic test results before deciding to insure you? What about employers, before hiring you?
Where are you protected from genetic discrimination?
Different regions have different legislation and regulation of genetic testing.
In March 2017, Canada passed Bill S-201, the Genetic Non-Discrimination Act.
The Act prevents people being pressured to undergo genetic testing in order to be eligible for goods or services (such as insurance); or from having to disclose their results.
It prohibits employers from discriminating against workers on the basis of any genetic test results. It also amends the Canadian Human Rights Act to prohibit discrimination based on genetic characteristics.
The Canadian Coalition for Genetic Fairness is made up of several advocacy groups, such as the Parkinson Society of Canada or Muscular Dystrophy Canada. They also promote genetic nondiscrimination.
In the United States, the Genetic Information Nondiscrimination Act of 2008 (GINA) prohibits discrimination based on genetic information in health insurance and employment. Health insurance companies are not allowed to deny coverage to healthy people based solely on potential genetic risk, nor are employers allowed to discriminate based on genetic data.
In the UK, the 2010 Equality Act prevents employers from discriminating based on genetic data.
In the European Union countries, the 2010 Lisbon Treaty prohibits discrimination based on “genetic features”.
The United Nations Educational, Scientific and Cultural Organization (UNESCO) adopted the Universal Declaration on the Human Genome and Human Rights in 2003 and International Declaration on Human Genetic Data in 2012, which includes provisions for preventing genetic discrimination and any use of genetic information that would contravene dignity, freedom, and human rights.
What behavioral factors are involved?
Once you get your test results, what could you do? What should you do?
What are you willing to change, or not change?
Who can help you understand all the information?
What moral and ethical questions are involved?
Are you fully informed about the test, and what it involves, before you do it (a.k.a. “informed consent”)?
Are you obligated to do anything once you know your results?
For instance, if you’re young enough and considering having a family, should the results of genetic testing change your choice to reproduce?
What if you discover some awkward truths about your genetic heritage or parentage?
Can, and should, we patent genes?
Fun factoid!
In one key case (Association for Molecular Pathology v. Myriad Genetics, No. 12-398 [569 U.S.June 13, 2013]), Judge Robert Sweet found that the claims on DNA molecules were invalid because “DNA represents the physical embodiment of biological information, distinct in its essential characteristics from any other chemical found in nature”.
We won’t always have the “right” answers to any of these questions.
But we should be asking them.
What’s up next: Exploring specific tests
In this chapter, we’ve given you a broad background to consider.
In the next chapter, we’ll look at specific types of tests, and what we discovered about them with our own testing practices.





